Single Cell and Spatial Transcriptomics
Unlock the Power of Single Cell and Spatial Transcriptomics for Unprecedented Biological Insights
Single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics are revolutionary techniques that offer unparalleled insights into cellular function and tissue architecture. scRNA-seq allows researchers to analyze the gene expression profiles of individual cells, uncovering cellular heterogeneity and revealing intricate biological processes at a resolution previously unattainable. Spatial transcriptomics complements this by preserving the spatial context of gene expression within tissue samples, enabling the study of gene activity in relation to tissue structure. Together, these technologies provide a comprehensive understanding of cellular dynamics and interactions, driving advancements in fields such as developmental biology, oncology, and personalized medicine.
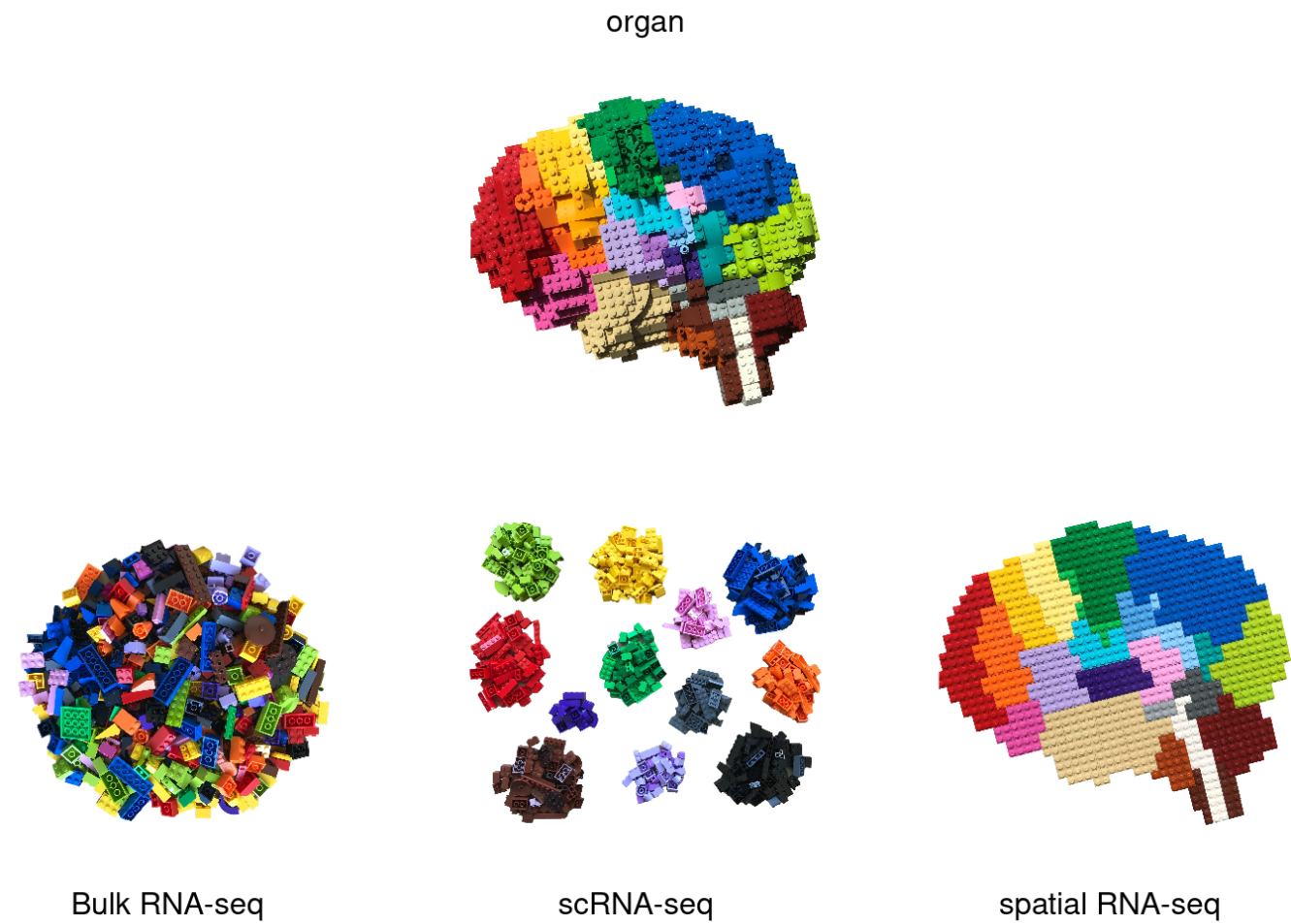
Image credit: @BoXia7
Single Cell Services
At the GCTU, we offer two leading single cell workflows - 10X Genomics and Parse Biosciences. Whilst both provide high quality single cell RNAseq data, we may recommend one workflow over the other depending on a number of parameters such as number of cells, cell population size, current sample status etc Please reach out to us at genomics@qub.ac.uk to discuss your single cell project and optimal workflow.
We accept single cell suspensions only - users will need to prepare these themselves before sending them to the unit at a pre-arranged delivery day and time.
10X Genomics
10X Genomics single-cell RNA-seq workflows are designed to analyze gene expression at the single-cell level. The process begins with cell isolation and barcoding using the Chromium X instrument, where each cell's RNA is captured and labeled with a unique barcode. Downstream sequencing libraries are prepared from the cell barcoded RNA and subjected to standard sequencing methods. Sequencing reads are then processed using the Cell Ranger pipeline, which includes steps like alignment to a reference genome, UMI counting for gene expression quantification, and identification of true cells versus background. The resulting gene expression matrix is used for downstream analysis, including cell clustering, visualization, and differential expression, providing detailed insights into cellular heterogeneity.
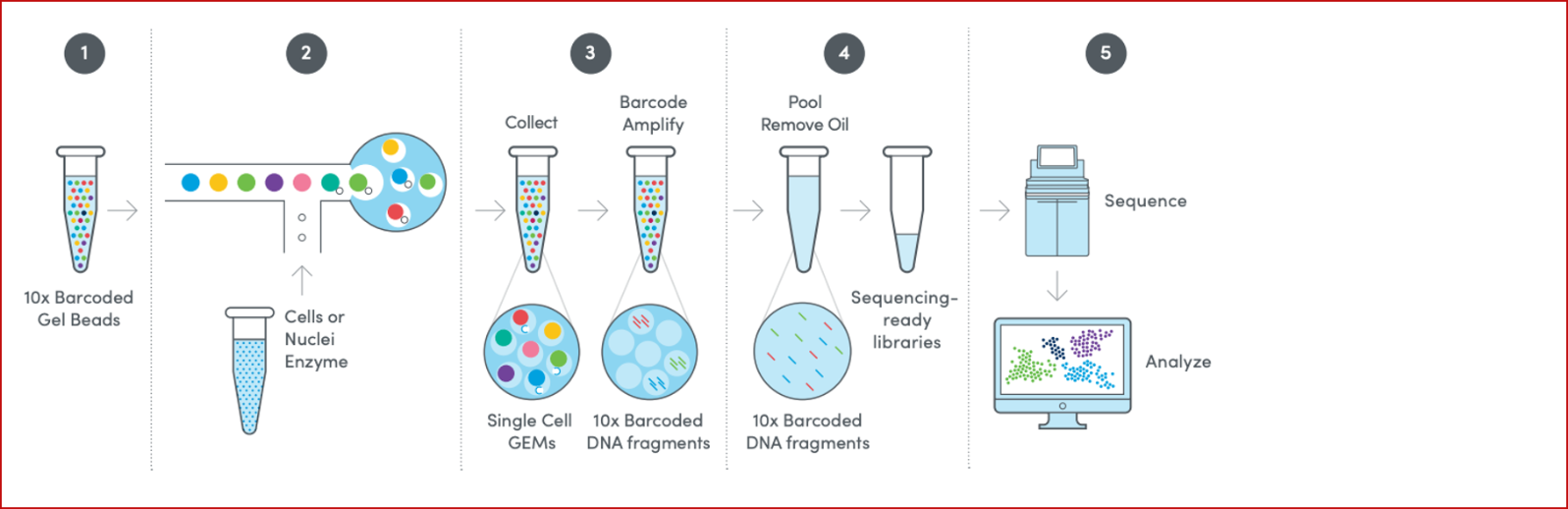
The GCTU offer the below 10X Genomics workflows:
- Chromium GEM-X Single Cell 3' Kit
- Chromium Next GEM Single Cell ATAC Kit
- Chromium Next GEM Single Cell Multiome ATAC + Gene Expression
- Chromium GEM-X Single Cell 5' Kit
Parse Biosciences
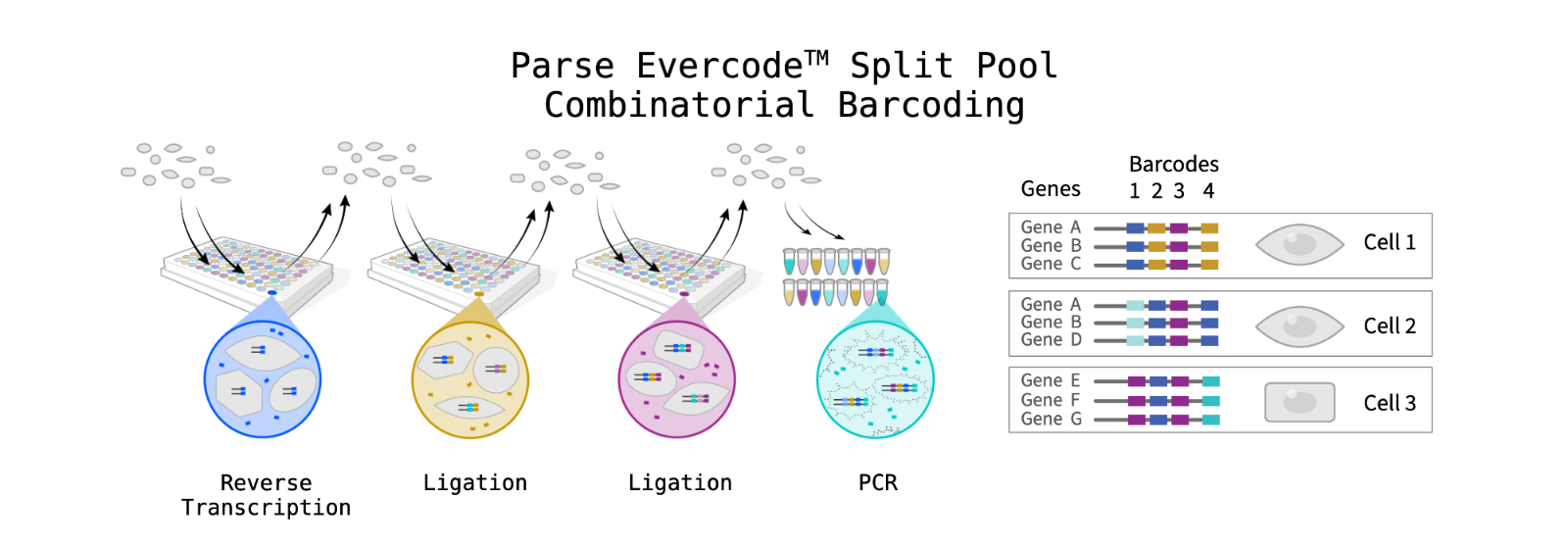
The GCTU offer the below Parse Bioscience workflows:
- Parse Evercode Mini Kit V3 (1-12 samples/10,000 cells)
- Parse Evercode WT Kit V3 (1-48 samples/100,000 cells)
- Parse Evercode Mega Kit V3 (1-96 samples/1M cells)
Spatial Transcriptomic Services
At the GCTU, we offer 10X Genomics spatial transcriptomic workflows - namely Visium, VisiumHD and Xenium. These complementary workflows differ in terms of number of genes detected, capture area size, sequencing requirements and available species. Please reach out to us at genomics@qub.ac.uk to discuss your spatial transcriptomic project and optimal workflow.
For Visium & VisiumHD we accept freshly cut tissue section on glass slides, whereas for Xenium we will provider users with specific Xenium slides on to which tissues are to be placed onto directly. We can also support tumour microarray (TMA) creation and use on both workflow - please reach out to discuss.
Visium & VisiumHD
The 10x Genomics Visium CytAssist workflow is tailored for spatial transcriptomics, enabling gene expression mapping within tissue sections while preserving spatial information. The Visium workflow affords researcher whole transcriptome information at multi-Cell (standard) or near single cell (HD) resolution. The workflow begins with the preparation of fresh-frozen or FFPE tissue sections, which are placed onto standard glass slides. The CytAssist instrument then transfers mRNA from the tissue sections to Visium slides with barcoded capture spots. These spots capture the mRNA while maintaining the tissue's spatial context. The captured RNA is reverse-transcribed into cDNA, which is then used to create sequencing libraries. After sequencing, the data is processed to generate spatial gene expression maps, providing insights into gene activity across different regions of the tissue, all while maintaining the original spatial architecture.
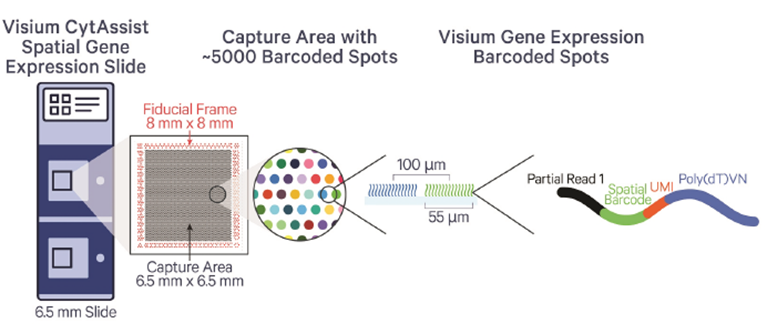
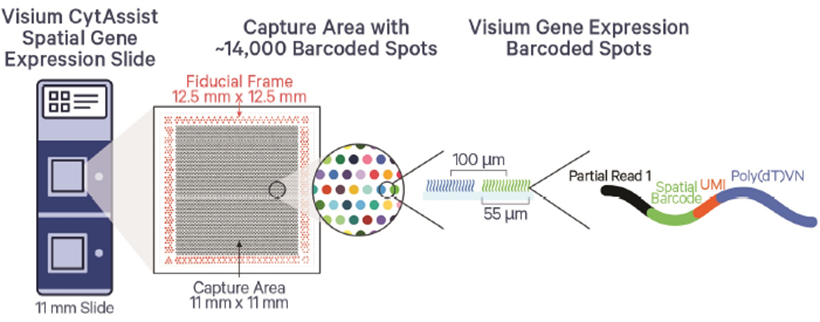
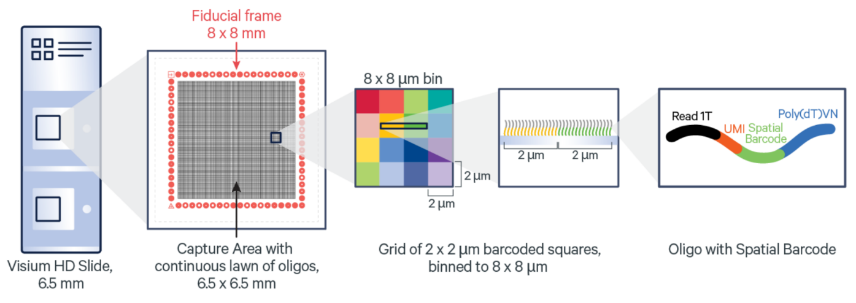
The GCTU offer the below Visium workflows:
- Standard Visium: resolution around 55µM, 2 capture areas per slide at 6.5x6.5mm or 11x11mm
- VisiumHD: resolution 2µM (binned to 8µM), 2 capture areas per slide at 6.5x6.5mm
Xenium
The 10x Genomics Xenium workflow is designed for high-resolution spatial transcriptomics, allowing detailed mapping of gene expression within tissue sections. The Xenium workflow affords researcher access to information for between 100-5,000 genes at sub-cellular resolution. The workflow involves preparing fresh-frozen tissue sections, which are placed onto Xenium slides with high-density barcoded capture areas. These areas capture and preserve mRNA from specific locations in the tissue. The captured RNA is then reverse-transcribed into cDNA and used to create sequencing libraries. After sequencing, the data is analyzed to produce high-resolution spatial gene expression maps, providing in-depth insights into tissue architecture and cellular function at a finer spatial scale.
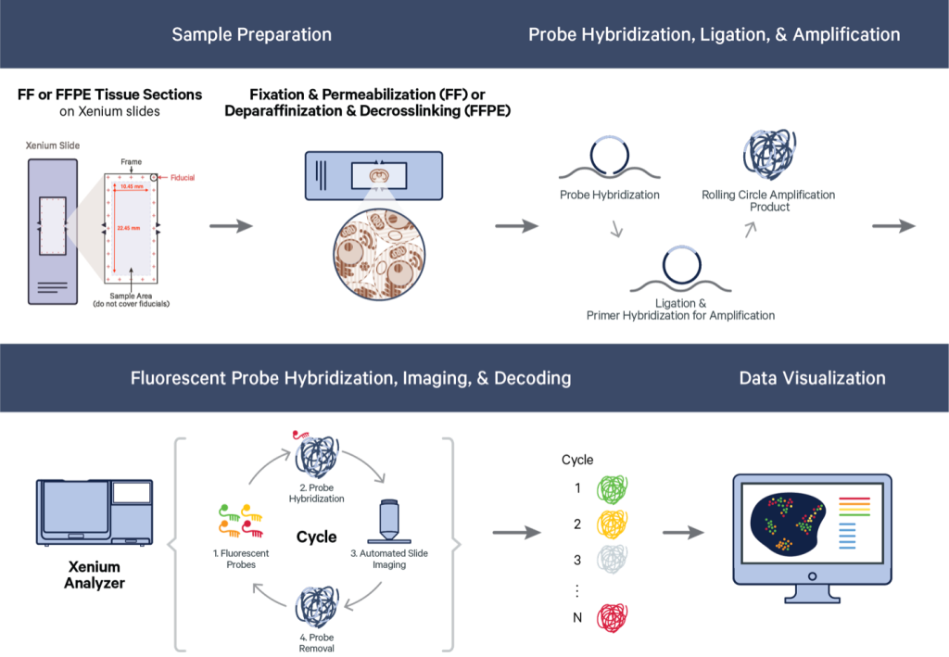
The GCTU offer the below Xenium workflow:
- Xenium run: sub-cellular resolution, 1 capture area per slide at 10.45mm x 22.45mm. Note - 2 slides must be run per Xenium instrument run.
- 10X Genomics offers a range of off the shelf panels for Human and Mouse, as well as fully customisable panels. See here for more information.